
Bioinformatics group
The Bioinformatics group uses computational methods to analyse genome sequences, amino acid sequences, and gene expression data, both to identify new genes of interest and to determine their structure, function and role in the cell. Advanced computational tools are both being used and developed. The group is also creating databases and web sites with our tools and generated data. We are involved in many collaborative projects with different research groups.
Challenges
Huge amounts of molecular biology data is being generated from a range of different technologies. New technologies allow extensive sequencing to be carried out to analyse sequence variation, transcription, epigenetics and other phenomena. Complete genome sequences from more than a thousand organisms as well as data from large-scale protein structure determination projects are also publicly available. The main challenge in computational biology is to integrate and make sense of all of this data.
Projects
- Structural bioinformatics: Computational models of the 3D structure of proteins are created and studied in order to understand the molecular mechanisms of enzyme activities. How do mutations affect the structure and function of a protein? How have the genes evolved? Docking and molecular dynamics simulations are also used in our studies.
- Tools for sequence analysis: Extremely rapid implementations of the fundamental algorithm for local sequence alignment that exploits readily available parallel computing technology has been developed (PARALIGN, SWIPE). We are working to improve such tools further and to apply them in important cases, like genome assembly or accurate read mapping of data from deep sequencing.
Recent achievements
We have developed the fastest implementation of the important Smith-Waterman local sequence alignment algorithm (BMC Bioinformatics, 2011). We have also discovered an important mutation in the AP endonuclease in lab strains of S.pombe (DNA repair, 2011). Furthermore, we have characterized the role of specific domains in the protein structure of PCSK9 in degradation of the LDL receptor (J Lipid Res 2011; BBRC, 2011) and contributed to the characterization of a PSC susceptibility locus (Nat Genet 2011).
Publications
Group leader
Centre for Molecular Biology and Neuroscience
Department of Microbiology
Oslo University Hospital
Rikshospitalet
PO Box 4950 Nydalen
NO-0424 Oslo, Norway
Mob: +47 90755587
E-mail: torognes@ifi.uio.no
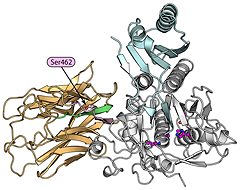
Structural model of the PCSK9 protein that mediates degradation of the low density lipoprotein (LDL) receptors. The important Ser462 residue that reduces secretion when mutated is indicated. From Cameron et al. (2009).
Latest 3 publications
Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
PLoS Genet, 9 (2), e1003260
PubMed 23408903
The human homolog of Escherichia coli endonuclease V is a nucleolar protein with affinity for branched DNA structures
PLoS One, 7 (11), e47466
PubMed 23139746
Alkbh1 and Tzfp repress a non-repeat piRNA cluster in pachytene spermatocytes
Nucleic Acids Res, 40 (21), 10950-63
PubMed 22965116
PO Box 1105 Blindern, NO-0317 Oslo, Norway. Tel: +47 22851528. Fax: +47 22851488
