
NeSys - Neural systems and graphics computing laboratory
Group home page at UiO
Group home page (old)
NeSys is a computational neuroanatomy and neuroinformatics laboratory. The research of the group focuses on 1) the development of new and powerful methods for computerized data acquisition, 3-D reconstruction, visualization and quantitative analyses of features in brain tissues, 2) data management and construction of 3-D brain atlases of experimental data, and 3) investigations on organization and re-organization of brain systems architecture in rat and mouse models, and 4) in vivo imaging in the context of multi-modality brain atlasing.
Challenges
Much of the research carried out today on rodent models generates high resolution image data, allowing characterization and analysis of brain molecular distribution, gene expression, and connectivity. It is of great importance not only to record more data but also to integrate data, re-use data in novel combinations, and perform more powerful analyses. To this end, data management systems and advanced analytical tools are needed. Structure and structure-function relationships are often better understood by introducing 3-D reconstruction and advanced visualization and modelling tools.
Projects
- Neuroscience databases and atlasing systems. We develop database applications for image data, from microscopy level to in vivo imaging data. We now host a rat and mouse brain work bench (www.rbwb.org), providing access to repositories, databases, and analytical tools, for circuit level as well as molecular distribution data.
- Localization in the brain. We develop and use technologies (robotic microscopy data acquisition, computerised 3-D reconstruction, and digital atlasing) for efficiently assigning localization to neuroscience data.
- Brain map transformations. We study design principles and changes in the architecture of major circuits in the brain following external and genetic manipulations.
- High resolution MRI and microPET. In several project collaborations, tomographical imaging techniques are employed to characterize structural and functional relationships occurring in the brain following experimental perturbations or disease.
Key achievements
- Development and sharing of software for 3-D reconstruction, visualization, and analysis of neuronal distribution and brain regional organization (www.rodentbrainworkbench.org).
- Development of a novel digital atlas system for evaluation of cellular distribution patterns in large series of high-resolution mosaic images of histological sections (Boy et al., NeuroImage 2006; 33: 449-462, www.rodentbrainworkbench.org).
- Mapping of topographical organization in cerebro-cerebellar pathways from somatosensory cortex through the pontine nuclei to different locations in the cerebellum (Odeh et al., J Neurosci. 2005; 25: 5680-5690; Leergaard et al., Eur J Neurosci. 2006; 24: 2801-2812).
- Establishment of the first database on Functional Anatomy of the Cerebro-Cerebellar System in rat (Bjaalie et al., Neuroscience 2005; 136:681-696; Moene et al., Neuroinformatics 2007, In Press; www.rodentbrainworkbench.org).
Group leader
 Professor
Jan G. Bjaalie
Professor
Jan G. Bjaalie
Department of Anatomy & CMBN
University of Oslo
PO Box 1105 Blindern
NO-0317 Oslo
Norway
Tel: +47 22851263 or +47 91787901
Fax: +47 22851045 or +47 22851278
E-mail: j.g.bjaalie@medisin.uio.no
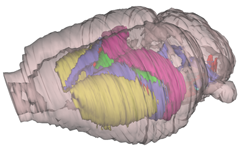
3-D digital rat brain model reconstructed from a standard stereotaxic atlas, serving as localization matrix in new database facilities.
PO Box 1105 Blindern, NO-0317 Oslo, Norway. Tel: +47 22851528. Fax: +47 22851488