News 2012
The Centre for Molecular Biology and Neuroscience (CMBN) has come to an end and this website will no longer be updated. For information about the research carried out in the research groups that comprised CMBN, please see the websites of UiO and OUS as well as the websites of the individual groups. Best wishes for 2013 to our collaborators and all visitors of the website! |
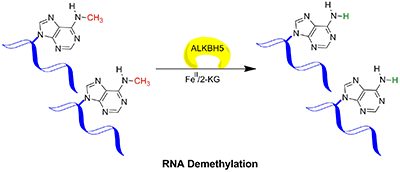
Methylation of mammalian DNA and histone residues are known to regulate transcription, and the discovery of demethylases that remove methylation in DNA and histones provide a basis for the understanding of dynamic regulation of mammalian gene expression. Knowledge on these demethylases has led to a tremendous progress in the understanding of methyl marks in gene regulation and role in numerous diseases. In mRNA, the methylation of adenosine (6meA) is particularly interesting since it is the most abundant internal modification. The first mRNA demethylase, FTO, was identified recently and 6meA was shown to be a substrate for FTO. Genome-wide association studies have identified a firm link between the human FTO gene, obesity and type II diabetes. In a collaborative study, Dahl (Picture) and colleagues, together with collaborators at the University of Chicago and Beijing, show that ALKBH5 is a second demethylase for 6meA in mRNA (see figure below) and that mice lacking this demethylase are infertile. Together, the discovery of two proteins that can reverse 6meA modifications from mRNA draws attention to the potential regulatory functions of reversible RNA methylation and the role of 6meA in disease. Insights into the mechanism of this process may well open up new horizons and opportunities for basic as well as translational research. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG and He C |
The article shows that doses of anesthesia insufficient to affect neuronal responses to whisker stimulation suppressed astrocyte calcium signals. Thrane AS, Thrane VR, Zeppenfeld D, Lou N, Xu Q, Nagelhus EA, Nedergaard M |
 |
The CMBN 10 year summary report is now available. |
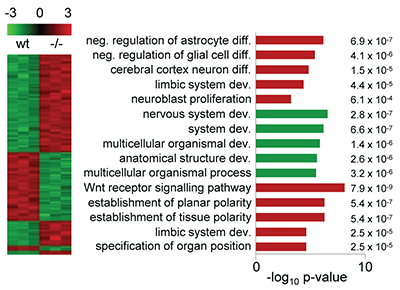
AlkB homolog 1 (ALKBH1) is one of nine members of the family of mammalian AlkB homologs. Most Alkbh1 deficient mice die during embryonic development, and survivors are characterized by defects in tissues originating from the ectodermal lineage. In this study, we show that ALKBH1 is a histone dioxygenase that acts specifically on histone H2A. Further, we demonstrate that deletion of Alkbh1 in embryonic stem cells leads to upregulation of the core genes involved in pluripotency and that ALKBH1 is required during early differentiation. Our results suggest that ALKBH1 is involved in neural development by modifying the methylation status of histone H2A. Ougland R, Lando D, Jonson I, Dahl JA, Moen MN, Nordstrand LM, Rognes T, Lee JT, Klungland A, Kouzarides T, Larsen E |
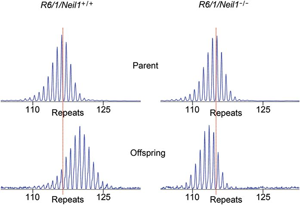
Two CMBN groups have identified a DNA repair gene that modify somatic and germline CAG trinucleotide repeat instability in the Huntingtin gene. Huntington's disease (HD) is a progressive neurodegenerative disorder caused by a CAG:CTG repeat expansion in exon 1 of the gene that encodes the polyglutamine-containing protein Huntingtin. It is shown here that somatic CAG expansions are significantly reduced in several organs of R6/1 mice lacking exon 2 of Nei-like 1 (Neil1). This study further confirm a role of oxidative DNA damage and neurodegeneration. We previously identified (with Cynthia McMurray's group at the Mayo Clinic, Rochester, MA, USA) a role of 8-oxoguanine-DNA glycosylase (OGG1) enzyme in initiation of age-dependent CAG trinucleotide expansion associated with Huntington's disease that occurs in somatic cells. (Kovtun et al., Nature 2007). Møllersen L, Rowe AD, Illuzzi JL, Hildrestrand GA, Gerhold KJ, Tveterås L, Bjølgerud A, Wilson DM, Bjørås M, Klungland A |
 |
CMBN scientists have co-authored the cover story of the current issue of Science Translational Medicine. The article describes paravascular pathways that others call the superhighways or hidden sewers of the brain. Iliff JJ, Wang M, Liao Y, Plogg BA, Peng W, Gundersen GA, Benveniste H, Vates GE, Deane R, Goldman SA, Nagelhus EA, Nedergaard M The article has been featured in several news stories: Vannkanaler renser hjernen Forklarer hjernens avfallsmekanisme Vannkanaler holder hjernen ren |
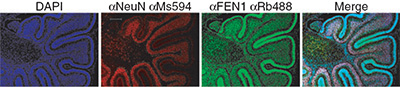
The structure specific flap endonuclease 1 (FEN1) plays an essential role in long-patch base excision 15 repair (BER) and in DNA replication. Here, the monitoring of the localization and kinetics of endogenous FEN1 is achieved by tagging the Fen1 gene in embryonic stem cells for generating "knockin" mice. In line with the role of FEN1 in processing of Okazaki fragments during DNA replication, FEN1-YFP expression is mainly observed in highly proliferating tissue. In vivo multi-photon fluorescence microscopy demonstrates rapid localization of FEN1 to local laser-induced DNA damage sites in nuclei, providing evidence of a highly mobile protein that accumulates fast at DNA lesion sites with high turnover rate. Inhibition of poly (ADP-ribose) polymerase 1 (PARP1) disrupts FEN1 accumulation at sites of DNA damage, indicating that PARP1 is required for FEN1 recruitment to DNA repair intermediates in BER.
Immunofluorescence staining of endogenous FEN1 (green) and brain cell marker anti-NeuN of Fen1y/y mouse brain 7.5 days after birth. DAPI (blue) stained nuclear DNA. To the right, a merge of blue, red and green channel is shown. The scale bar is 200 μm. Kleppa L, Mari PO, Larsen E, Lien GF, Godon C, Theil AF, Nesse GJ, Wiksen H, Vermeulen W, Giglia-Mari G, Klungland A |
Acute promyelocytic leukemia (APL) represents one of the most malignant forms of acute myelogenous leukemia. Despite the aggressiveness of this cancer, it can be effectively cured by two different drugs called all-trans retinoic acid (ATRA) and arsenic trioxide (ATO). In an article recently published in the journal Blood the authors describe a novel therapeutic mechanism that explains why ATO is particularely effective in curing APL. These findings may provide important clues to how the ATO-based therapy of APL may be further developed and adapted for treatment of other cancers. Lång E, Grudic A, Pankiv S, Bruserud O, Simonsen A, Bjerkvig R, Bjørås M, Bøe SO |
Cell proliferation during normal growth and development of eukaryotic organisms is regulated by cyclin dependent kinases (CDKs). CDKs are the master regulators of the cell cycle, which is the mechanism that controls cell duplication. Cancer cells deceitfully hijack CDKs to drive their uncontrolled cell proliferation. It is therefore very important to understand the function and regulation of CDKs. A team of researchers at CMBN has now discovered a new function for Cdk1 in the model organism budding yeast (Cdk1, also known as Cdc28, is the critical cell cycle regulator in yeast). Chymkowitch P, Eldholm V, Lorenz S, Zimmermann C, Lindvall JM, Bjørås M, Meza-Zepeda LA, Enserink JM (2012) |
In a collaborative study with scientists led by Prof. Leona Samson at MIT, Boston, it is shown that three genes collaborate to protect against inflammation-mediated colon carcinogenesis. More than 15% of cancer deaths worldwide are associated with underlying infections or inflammatory conditions and inflamed tissues harbor elevated etheno-base (ε-base) DNA lesions. The alkyl adenine DNA glycosylase (AAG) recognizes such lesions and provide some protecting against inflammation associated colon cancer. Two other DNA repair enzymes repair ε-base lesions, ALKBH2 and ALKBH3, and could also provide protection against inflammation-mediated colon carcinogenesis. Using established chemically induced colitis and colon cancer models in mice, it is shown here that Alkbh2 and Alkbh3 provide cancer protection similar to that of the DNA glycosylase Aag. Moreover, Alkbh2 and Alkbh3 each display apparent epistasis with Aag. Surprisingly, deficiency in all 3 DNA repair enzymes confers a massively synergistic phenotype, such that animals lacking all 3 DNA repair enzymes cannot survive even a single bout of chemically induced colitis. Calvo JA, Meira LB, Lee CY, Moroski-Erkul CA, Abolhassani N, Taghizadeh K, Eichinger LW, Muthupalani S, Nordstrand LM, Klungland A, Samson LD |
Adam Robertson, John Arne Dahl and colleagues at CMBN have published a protocol for identification of the novel 6th DNA base in genomic DNA. The base, 5-hydroxymethylCytosine (5-hmC), was discovered over 30 years ago. At that time, the 5-hmC modification was suggested to be a rare and nonmutagenic DNA damage lesion and therefore was given little attention. In early 2009, 5-hmC was identified again; however, at that time the importance of 5-hmC in epigenetics was realized, as two independent groups began the initial characterization of the 5-hmC modification. Although the function of the 5-hmC modification remains unclear, it has become clear that identifying genomic regions that contain 5-hmC will help to elucidate the function of this base. The protocol described here is based on the procedure developed to isolate 5-hmC–containing DNA in two steps (i) glucosylation of 5-hmC and (ii) the subsequent pull-down of β-glu-5-hmC by JBP1-coated magnetic beads. |
Scientists in CIR and CMBN published a paper in Nature Communication that reveals the intermolecular interactions of the FcRn-albumin complex. Albumin is the most abundant protein in blood where it has a pivotal role as a transporter of fatty acids and drugs. Like IgG, albumin has long serum half-life, protected from degradation by pH-dependent recycling mediated by interaction with the neonatal Fc receptor, FcRn. Although the FcRn interaction with IgG is well characterized at the atomic level, its interaction with albumin is not. Here we present structure-based modelling of the FcRn–albumin complex, supported by binding analysis of site-specific mutants, providing mechanistic evidence for the presence of pH-sensitive ionic networks at the interaction interface. These networks involve conserved histidines in both FcRn and albumin domain III. Histidines also contribute to intramolecular interactions that stabilize the otherwise flexible loops at both the interacting surfaces. These results may guide the development of novel albumin variants with altered serum half-life as carriers of drugs. Andersen JT, Dalhus B, Cameron J, Daba MB, Plumridge A, Evans L, Brennan SO, Gunnarsen KS, Bjørås M, Sleep D, Sandlie I (2012) |
PO Box 1105 Blindern, NO-0317 Oslo, Norway. Tel: +47 22851528. Fax: +47 22851488